Ribocentre Aptamer - Help
About Ribocentre-aptamer database
Ribocentre-aptamer Database is designed to provide information on RNA aptamers. Each aptamer page includes a brief introduction and offers various search options, such as sequence, publications, structures, binding affinities, and applications. Users can submit new RNA aptamer cases or related comments through the submission portal to enhance our database. The Binding Sites page compiles information on binding sites verified by published studies and classifies the aptamers. The Applications page showcases examples of RNA aptamer applications in previous research, updated regularly. The Structures page offers detailed insights into different aspects of RNA aptamers, and the Publications page provides links to individual aptamer pages, structural details, and related publications.
How to use the database
Search channels
On the home page, we start with a brief introduction, hoping that you will understand some basic concepts of RNA aptamers. There are three available search channels on the home page: (1) You can use the navigation bar to switch to the Aptamers, Applications, or Fluorescence pages. (2) You can search by keyword in the search box in the header. (3) Above the table, you can search by keyword or switch to a specific page.
Advanced search functionality
For a more precise search experience, you can visit the dedicated search page. On the search page, you can use multiple filtering criteria to narrow down your search scope, including but not limited to aptamer types, binding affinity ranges, publication years, and more. This advanced search functionality helps you quickly find RNA aptamer information that meets specific requirements, improving search efficiency and accuracy.
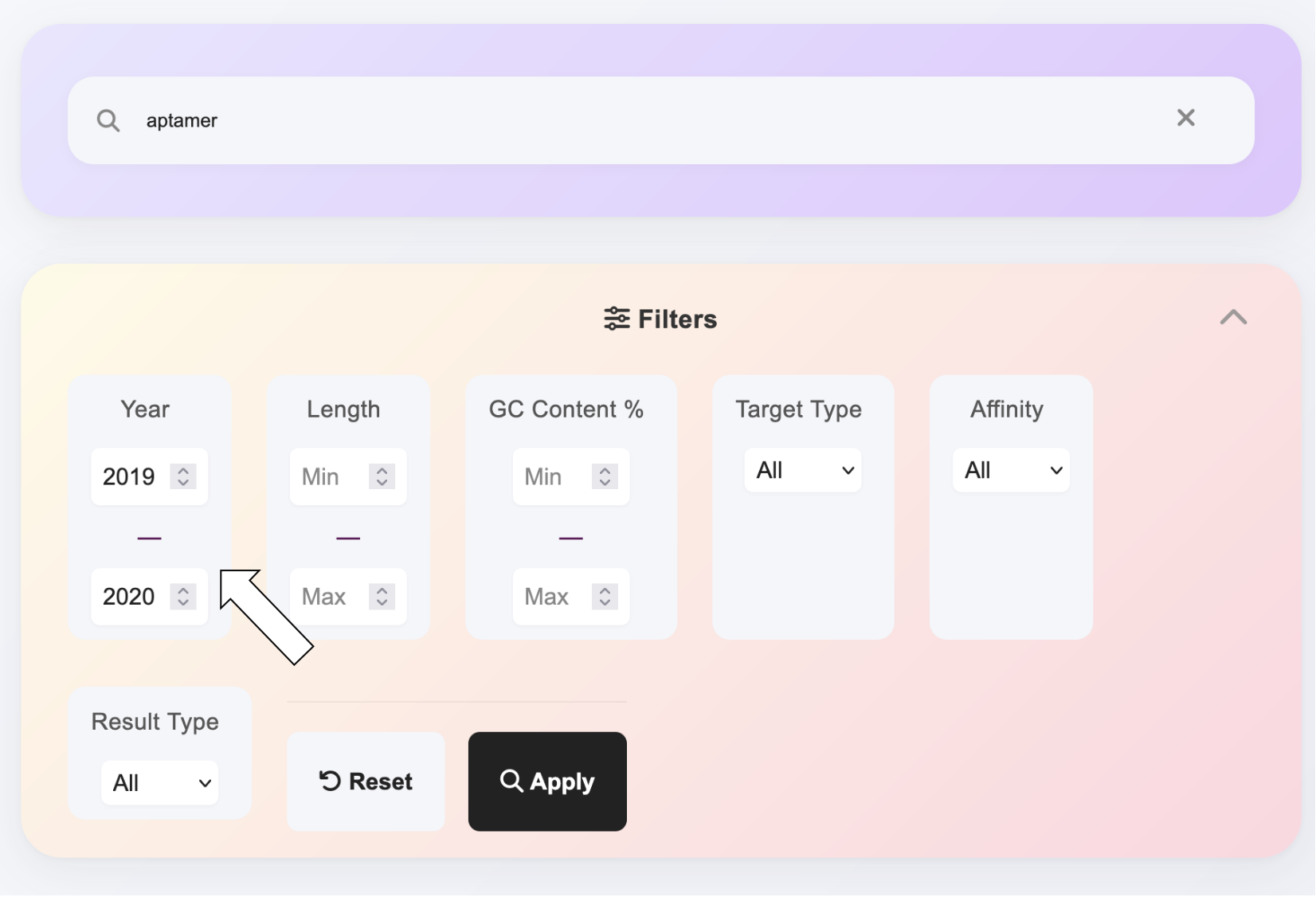
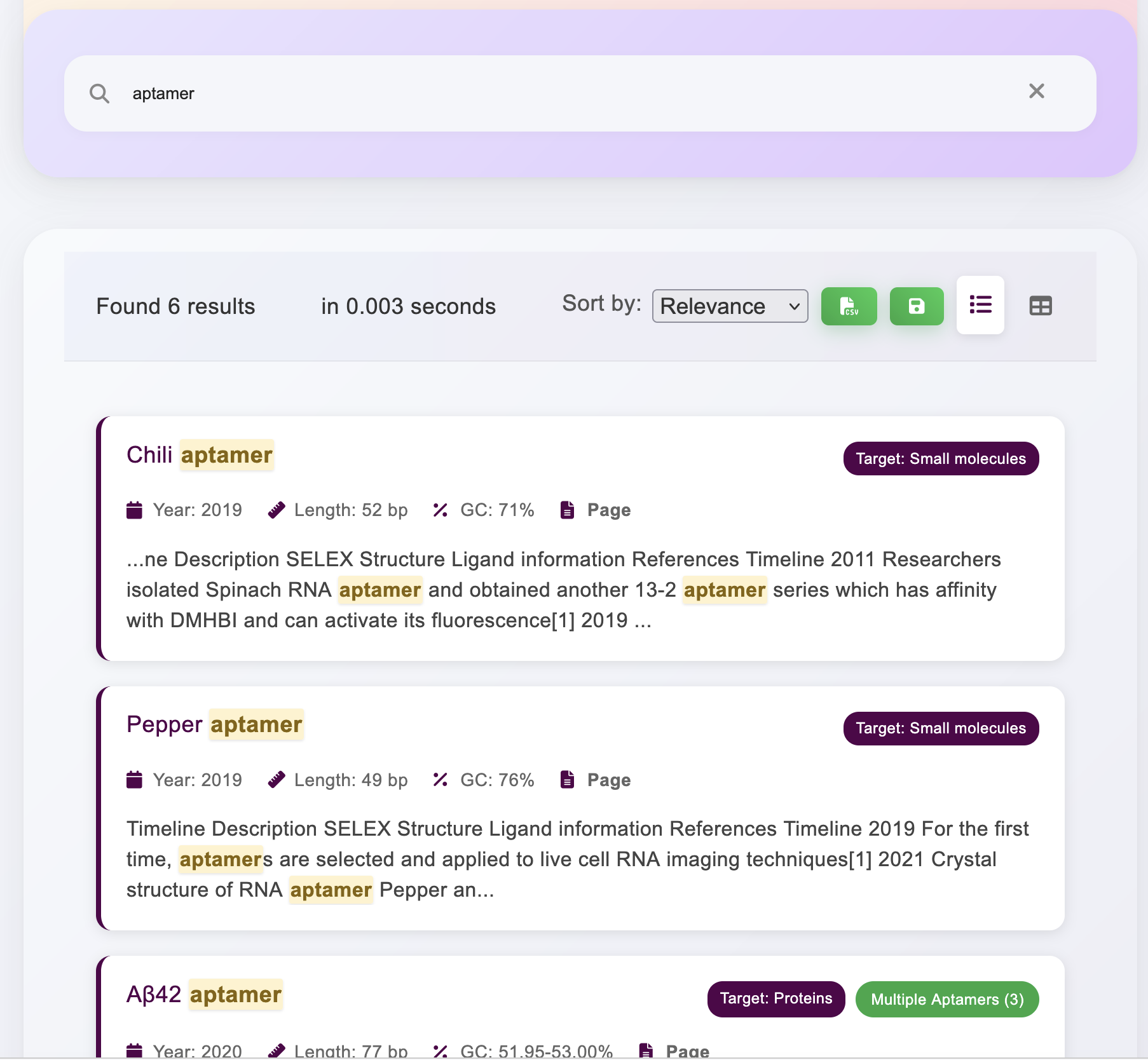
Download channels
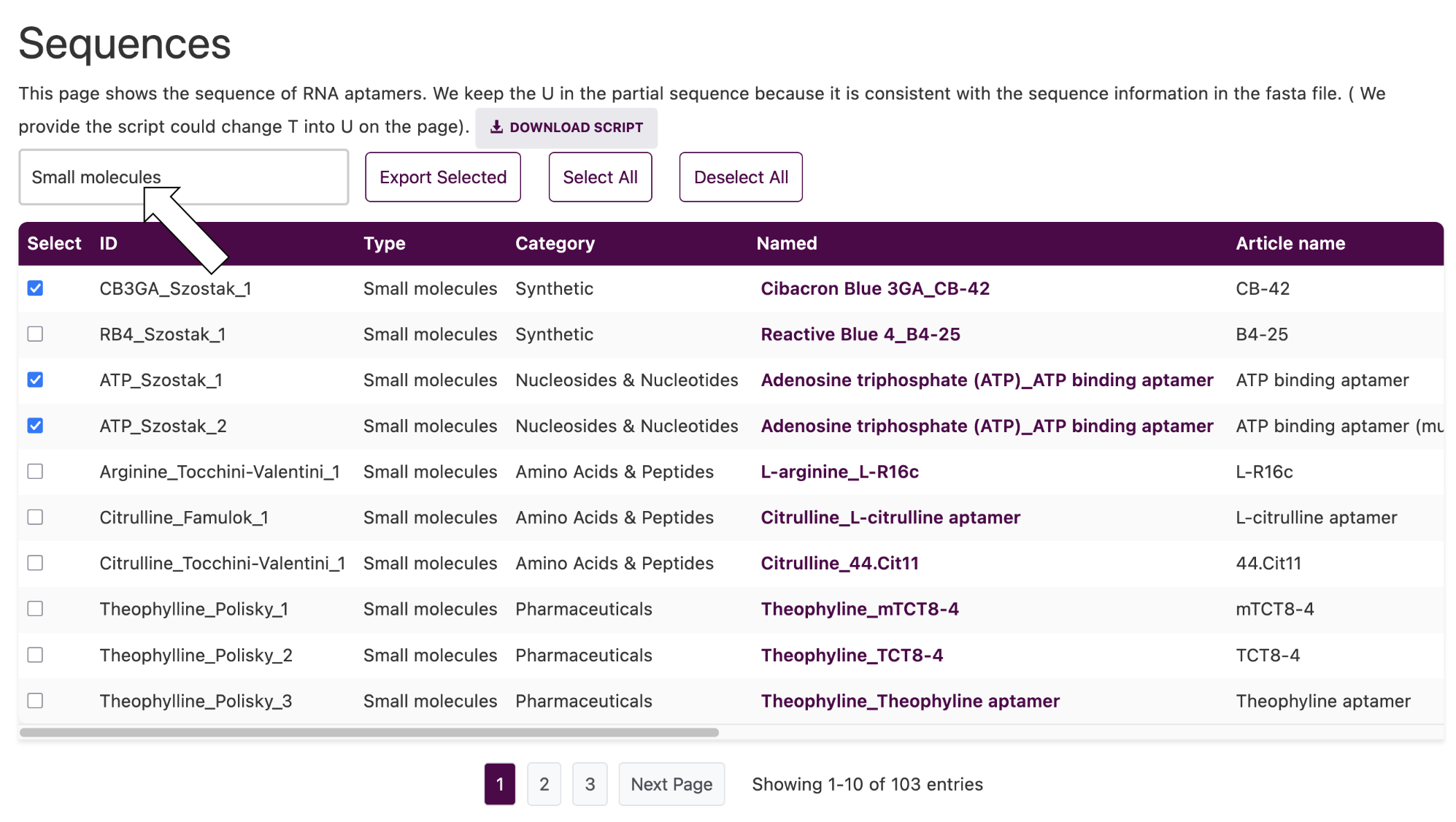
In each page, we mainly display the information of different RNA aptamers in table form. You can click the download button to download the information in different formats. When you want to download the information of a specific aptamer, you can filter it by search first and then download it.
Interactive visualization
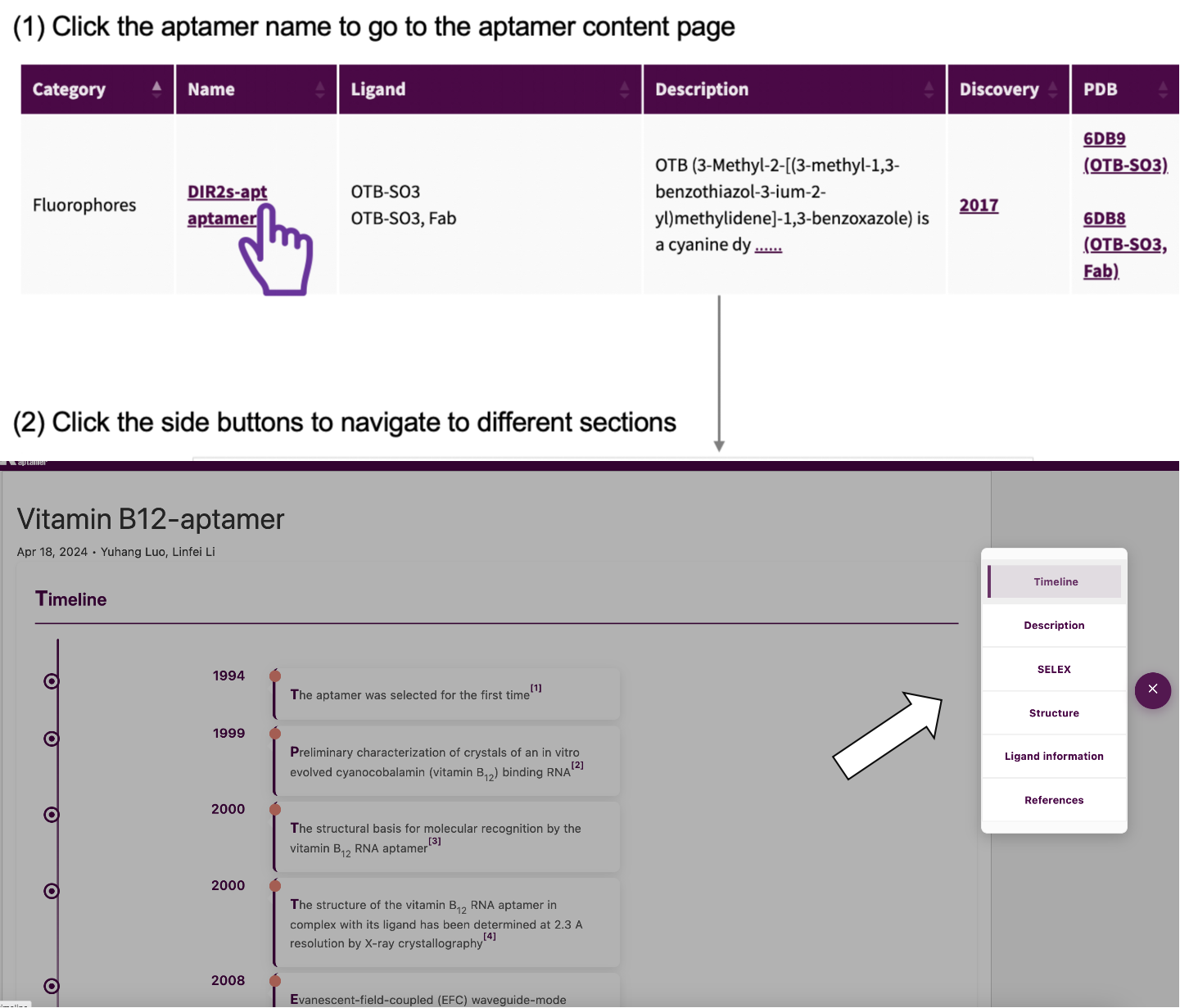
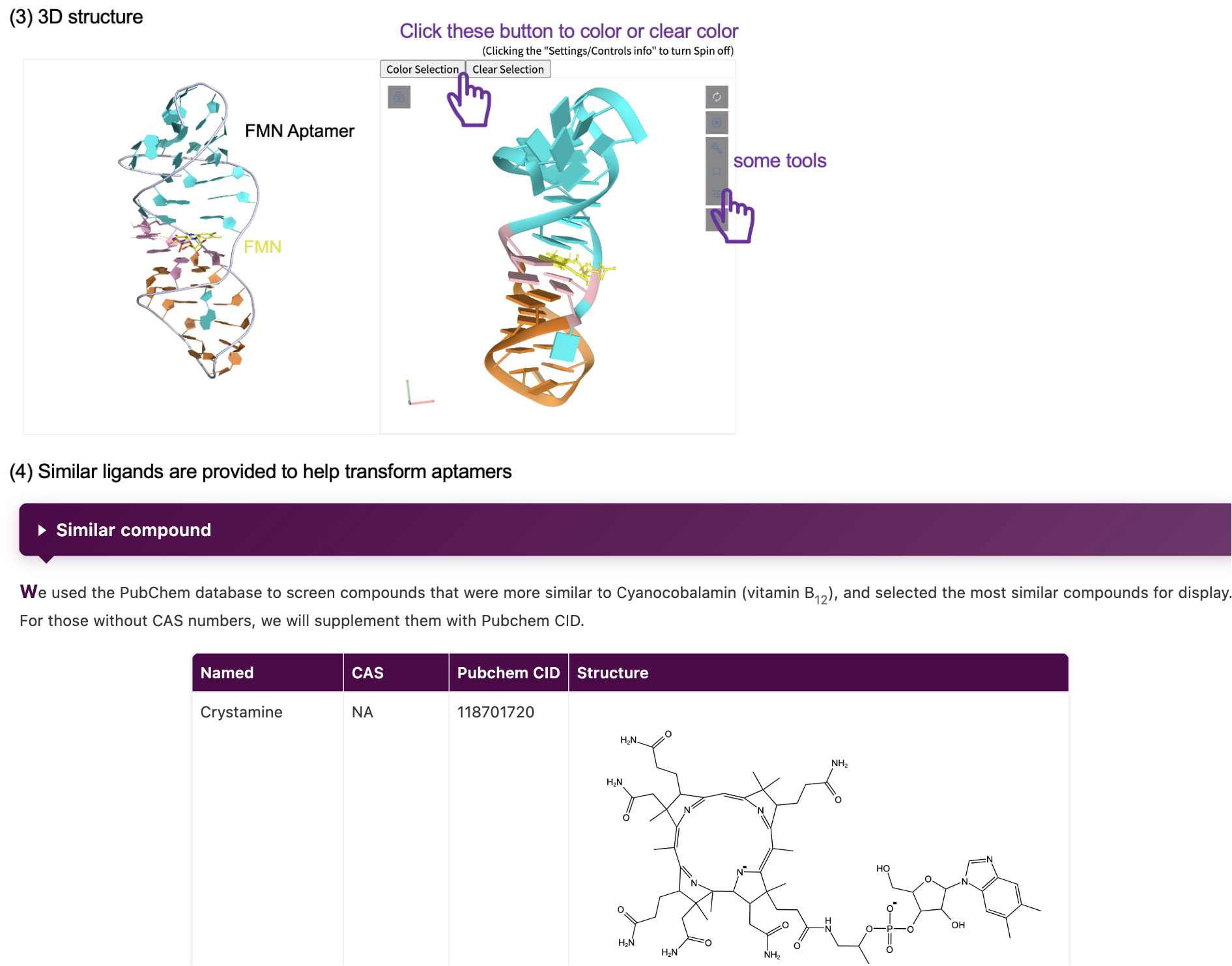
Clink the aptamer name in aptamer page, and it would send you to a new page which shows the timeline, structure, binding pockets information and so on of the riboswitch you choosed. For 3d structures you can use the interactive window to colour the structure or click on the zoom button on the right to go to a new page with more detailed information. In addition, we provide information about similar ligands to help you transform aptamers. We hope you can find all the information available in this database.
Feedback channel
To keep pace with the latest developments in RNA aptamers research, the database will undergo regular updates. In home page, We provide a link to the feedback button. In addition, Click the top button to return to a series of database collections.
API Access
The Ribocentre-Aptamer platform provides a robust API for programmatic access to our aptamer database. This API enables researchers to integrate our data into automated workflows, perform batch queries, and access comprehensive aptamer information in a structured format. Both web interface and programmatic access support URL-based search parameters for real-time data filtering.
API Endpoints
Our API offers multiple access points for retrieving aptamer data:
- Sequences Page:
https://aptamer.ribocentre.org/sequences/- Interactive web interface with search support; returns filtered HTML by default, JSON withformat=jsonparameter - JSON API:
https://aptamer.ribocentre.org/api/- Dedicated API endpoint, always returns JSON format with enhanced debugging information
Query Parameters
The API supports various query parameters for filtering and searching:
- search: Search across all aptamer fields (sequence, ligand, name, etc.)
- id: Filter results by specific aptamer ID
- category: Filter by aptamer category
- type: Filter by aptamer type (RNA, DNA)
- format: Request data in JSON format (only for
/sequences/endpoint) - limit: Limit the number of results returned (only for
/api/endpoint) - offset: Specify pagination offset (only for
/api/endpoint)
Usage Examples
Here are some common API usage patterns:
# Search for ATP-related aptamers (JSON API - always returns JSON) curl "https://aptamer.ribocentre.org/api/?search=ATP" # Get specific aptamer by ID (JSON API) curl "https://aptamer.ribocentre.org/api/?id=ATP_Szostak_1" # Get results with pagination (JSON API supports limit/offset) curl "https://aptamer.ribocentre.org/api/?search=DNA&limit=10&offset=0" # Search via sequences page with JSON format curl "https://aptamer.ribocentre.org/sequences/?search=thrombin&format=json" # Search via sequences page (returns filtered HTML results) curl "https://aptamer.ribocentre.org/sequences/?search=thrombin" # Visit sequences page with search results in browser # https://aptamer.ribocentre.org/sequences/?search=thrombin
Response Format
API responses include comprehensive debugging information and statistics:
{
"success": true,
"message": "Found 5 result(s)",
"query": {
"search": "ATP",
"timestamp": "2025-01-01T12:00:00.000Z"
},
"statistics": {
"total_in_database": 250,
"filtered_results": 5,
"returned_results": 5
},
"data": [...]
}
Data Fields
Each aptamer entry contains comprehensive information including sequence, target ligand, binding affinity, structure details, and publication references. The API provides access to all fields available in the web interface, enabling complete data integration.
Best Practices
For optimal API usage, we recommend:
- Implement reasonable delays between requests to avoid overwhelming the server
- Cache frequently accessed data locally to reduce server load
- Include proper error handling for network issues
- Always cite our database when using data in publications
For detailed API documentation and technical specifications, please visit our dedicated API Documentation page.
How to contact us
For any inquiries or concerns regarding the database, please reach out to 2112240208@gdpu.edu.cn.
Group Members
 Home
Home Database
Database Research
Research About us
About us




